A Master’s student class project can point us in the right direction.
Earlier this spring, plans across campus shifted as MIT led physical distancing initiatives to protect campus students, faculty, and staff. As part of this effort, all classes for the remainder of the semester have gone online and many courses, or students have altered their learning plans to accommodate the many new challenges stemming from a global pandemic.
Erika Madrian is a Master’s student in the Technology and Policy Program at MIT studying Infectious Disease Policy. She works in the MIT Biological Engineering Department where she studies RNA modifications in infectious bacteria. As part of her course requirement for the Humanitarian Supply Chain Lab’s MIT SCM.284, Erika turned her attention to the supply chain problems involved in COVID-19 diagnostic testing for her lab project.
Along with her advisor, Chelsey Graham (MIT SCM ’14), Erika determined her first step in the project was to make a map or checklist of all the components necessary to accomplish a q-PCR test for the virus (below). This resource list or what Professor Yossi Sheffi calls, “Exploding the BOM (Bill of Materials), serves as an insightful starting point to explore systems complexity. In Erika’s own words, below the checklist graphic is an abstract for this part of the project.
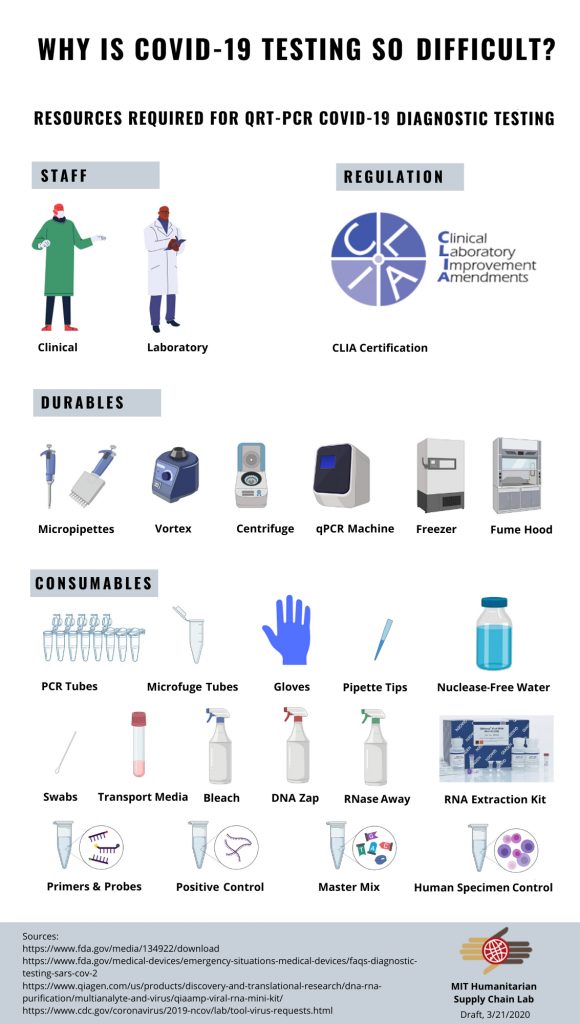
As Epidemiologist Larry Brilliant said in a recent interview*, it is possible to beat the novel coronavirus, but first we need a lot more testing. Since the first confirmed US COVID-19 case was identified on January 21 in Seattle, WA, the United States’ ability to rapidly test and accurately identify COVID-19 cases in the country has been less than adequate to meet demand. But why is testing so hard? Aside from strict regulatory issues mandated by the FDA initially, the testing supply chain and lab processes are complicated and involve many inputs, including lab consumables, durables, and proper laboratory staffing. In an attempt to better understand the major bottlenecks, the MIT Humanitarian Supply Chain lab has been researching the needed requirements to procure and perform q-PCR** tests for COVID-19. Even as rapid diagnostic tests are developed, qPCR will remain as the “gold standard” for testing. If just one of the resources from this checklist is missing, a qPCR COVID-19 test can’t be performed.
The COVID Diagnostic Resource Checklist is an example of a deliverable of an independent student project for Spring MIT SCM.284. While Erika continues her work in the course, we look forward seeing additional insights and discovery.
This project continues through May. It aims to develop a model that will identify bottlenecks in the supply chain of qPCR resources in order to scale testing capacity, which is essential to address the COVID-19 pandemic.
Download a pdf of COVID-19 Diagnostic Checklist
Details about the q-PCR test
Current COVID-19 testing looks for the presence of SARS-CoV-2 (the virus that causes COVID-19) genetic material in a patient’s sample through the following process:
First, the clinician collects a sample from the patient, typically by swabbing the nose or throat. The swab is placed in viral transport media, which preserves the integrity of the sample while it is sent to a diagnostic lab. Since SARS-CoV-2 stores its genetic material as RNA, the next step is to extract the RNA from the patient’s sample. The result will be mostly human RNA mixed with some viral RNA (if the patient does have COVID-19). There are many commercially available RNA extraction kits, though some require additional specialized equipment.
Finally, the isolated RNA is analyzed for the presence of SARS-CoV-2 genes using the qRT-PCR assay. This technique duplicates a select sequence of genetic code through a series of chemical reactions. “Primers” specify which sequence will be amplified, while a fluorescent probe signals each time the sequence has been duplicated. The reaction is carried out in a qPCR machine, which measures the fluorescent signal. While the CDC supplies primers and probes that detect sequences unique to SARS-CoV-2, it has also authorized the use of several other commercially available primers. If SARS-CoV-2 is present in the sample, the primers will recognize its genetic sequence and the probes will create a fluorescent signal, giving a positive test result for SARS-CoV-2.
Notes on the team
Erika Madrian is a Master’s student in the Technology and Policy Program at MIT studying Infectious Disease Policy. She works in the MIT Biological Engineering Department where she studies RNA modifications in infectious bacteria. She has a Bachelor’s degree from Tufts University in Biochemistry.
Chelsey Graham works at the MIT Center for Transportation and Logistics in the Humanitarian Supply Chain Lab. She has both private sector and humanitarian supply chain experience at Office Depot, Amazon, and the UN World Food Programme. Most recently she worked on the University of Washington’s Seattle Flu Study team, with a focus on rapid testing and intervention to detect and respond to pandemics. She holds a Master’s degree from MIT’s Supply Chain Management program.

Sources
*https://www.wired.com/story/coronavirus-interview-larry-brilliant-smallpox-epidemiologist/
**q-PCR (quantitative polymerase chain reaction) is a laboratory technique based on the polymerase chain reaction (PCR), which is a way of creating copies of DNA in a tube. PCR is a process thereby where DNA samples are amplified through a repeated heating process so that the sample is large enough to study.
Checklist Graphic Sources
https://www.fda.gov/media/134922/download
https://www.cdc.gov/coronavirus/2019-ncov/lab/tool-virus-requests.html
